Please follow us on Twitter for updates and news regarding SDMtoolbox!
October 26, 2023. Updated SDMtoolbox to version 2.6. I fixed an few major bugs introduced by Python updates that caused several functions to not work in ArcMap 10.8.
Please also note in recent years, many Antivirus softwares and Windows 11 are complicating the function of SDMtoolbox. For example, be sure to ‘unblock’ the MaxEnt software after downloading, else it will not work in Windows 11. Many antivirus softwares are increasingly perceiving SDMtoolbox as a potential virus. The reason for this is that SDMtoolbox is running java and python scripts in the background. If you are running into new and strange issues that are causing SDMtoolbox to no longer work. Try temporarily disabling antivirus and running the tool again. Be sure to turn antivirus back on. Also best to disconnect from internet while you are undefended.
October 28, 2021. Updated SDMtoolbox to version 2.5. I fixed a lot of minor bugs that I have been finding over the last year.
February 2, 2020. The beta of SDMtoolbox PRO is out! The creation of this version was a significant overhaul and required over 300 hrs of coding/troubleshooting. There certainly are additional bugs that I haven’t found, that will need fixing. Please be patient, but also let us know what your issues are. There remains a bit of work on this software, but since the software is 99% functional, I need to take a break from wrangling code and focus on other stuff. In a couple of months, I plan to come back to this to put the finishing polish on it and write a new user guide. Then I also will fix half a dozen issues in SDMtoolbox v2. Both versions will be fully maintained until most people transition to ArcGIS Pro (then SDMtoolbox v2 will be retired).
February 18, 2019. SDMtoolbox 2.4 is released and brings the use of multiple CPU cores to the ‘Run MaxEnt: Spatial Jackknifing’. Please update.
Have questions. Try SDMtoolbox’s forum!
SDMtoolbox is a python-based ArcGIS toolbox for spatial studies of ecology, evolution and genetics. SDMtoolbox consists of a series python scripts (92 and growing) designed to automate complicated ArcMap (ESRI) analyses. A large set of the tools were created to complement MaxEnt species distribution models (SDMs) or to improve the predictive performance of MaxEnt models (for an overview, see chapter 5 in the user guide Running a SDM in MaxEnt: from Start to Finish). MaxEnt uses maximum entropy to model species’ geographic distributions using presence-only data (Phillips et al. 2006) and has become one of the most prevalent methods due to its high predictive performance, computational efficiency and ease of use. SDMtoolbox is not limited to analyses of MaxEnt models and many tools are also available for use on other data (i.e. haplotype networks) or the results of other SDM methods (see Universal SDM Analyses).
Software Citations:
SDMtoolbox 2.0
Brown JL, Bennett JR, French CM (2017). SDMtoolbox 2.0: the next generation Python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. PeerJ PDF
SDMtoolbox PRO & SDMtoolbox 1.0
Brown J.L. (2014). SDMtoolbox: a python-based GIS toolkit for landscape genetic, biogeographic, and species distribution model analyses. Methods in Ecology and Evolution.
Brief Overview of Main Analyses:
- Calculation of species richness, weighted endemism and corrected weighted endemism
- Calculation of least-cost corridors and least-cost paths among shared haplotypes or among all sites (see image to right)
- Run spatial jackknifing in MaxEnt. Also automated independent evaluation of many feature classes and regularization multiplier values (see image to right)
- Creation of MaxEnt bias files for sampling biases associated with latitudinal changes in the area encompassed by decimal degree units
- Creation of MaxEnt bias files to limit background point selection to a maximum distance from presence points or within a buffered minimum-convex polygon (MCP) of a species’ distribution
- Spatially rarefy occurrence data (a.k.a. spatial filtering) to reduce spatial auto-correlation of occurrence points for use in species distribution modeling
- SDM over-prediction correction: clip by buffered MCP (see image to right)
- Limit dispersal in future SDMs
- Create a friction layer from a species distribution model (a.k.a. ecological niche model or environmental niche model)
- Calculate area of habitat contraction, expansion and other distribution changes between current and future SDMs (see image to right)
- Calculate vectors of core distributional changes between current and future SDMs
- Randomly select points
- Split shapefile by field attributes
- Explore summary statistics and correlations between environmental rasters before running a SDM
- Batch raster processing (i.e. preparing Worldclim data for MaxEnt): ASCII to raster files, raster to ASCII files, project to any projection, clip to a particular extent, re-sampling resolution, reclassifying and summing many rasters
- Split SDM by input clade relationship- Inverse Distance Weighting
- Create tessellated hexagon shapefiles
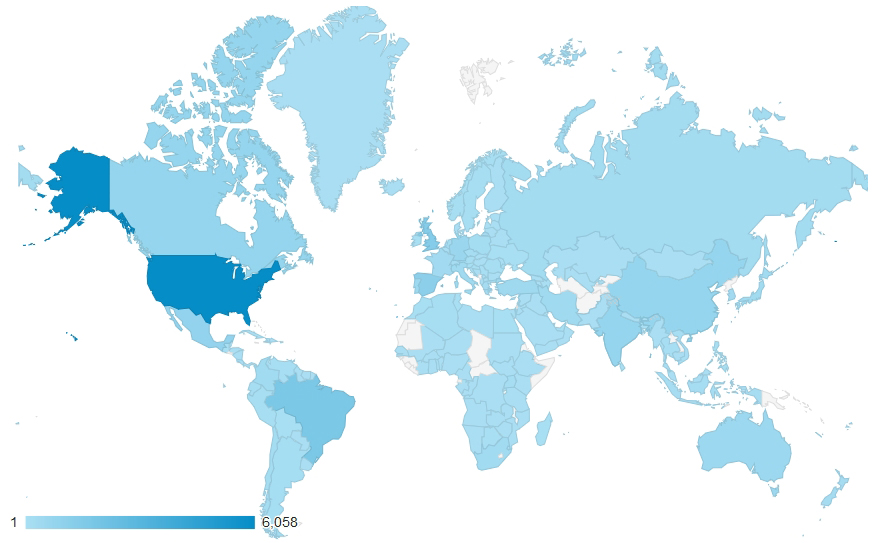
February 14, 2019. SDMtoolbox reaches 50,000+ users from all over the globe (map depicts downloads from 2012-2017)!